Research
Finding functional deubiquitinating enzymes
In Cancer Biology and Stem Cell Biology using CRISPR/Cas9 System
Deubiquitinating enzymes (DUBs) knockout library kit as a tool to screen functional DUBs for the target proteins
Dubbing Stemness-related Proteins and Onco-Proteins
Post-translational modifications (PTMs) of stemness-related proteins are essential for stem cell maintenance and differentiation. In stem cell self-renewal and differentiation, PTM of stemness-related proteins is tightly regulated because the modified proteins execute various stem cell fate choices. Ubiquitination and deubiquitination, which regulate protein turnover of several stemness related proteins, must be carefully coordinated to ensure optimal embryonic stem cell maintenance and differentiation. Deubiquitinating enzymes (DUBs), which specifically disassemble ubiquitin chains, are a central component in the ubiquitin proteasome pathway. These enzymes often control the balance between ubiquitination and deubiquitination. To maintain stemness and achieve efficient differentiation, the ubiquitination and deubiquitination molecular switches must operate in a balanced manner.
Similarly, many critical cellular events are under the control of this system, including those of cell division. It is therefore not surprising that a growing list of human cancers is associated with deregulated activity of these enzymes. Thus, we are specifically focusing on the involvement of DUBs in the stem cell biology, development of cancer, cell cycle, odontogenesis, and neurogenesis. To accomplish this, we are using cutting edge techniques such as CRISPR/Cas9 system, RNA interference, various protein analysis tools and advanced imaging techniques to study the mechanisms of deubiquitinating enzymes in cancer and stem cell biology.
Bio-Editing using CRISPR/Cas9 System
The most recent technology for genome editing is based on clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPRassociated protein 9 (Cas9), a defense system found in bacteria and archaea. The CRISPR/Cas9 technology has emerged as a potentially facile and advantageous alternative tool to ZFNs and TALENs for nuclease-mediated targeted genetic alterations. In the CRISPR/Cas9 system, a targeted site for DNA cleavage is guided by a short sequence of RNA to create the DSBs with high efficiency and specificity. We are using this tool to generate several disease models in human embryonic stem cells and induced pluripotent stem cells.
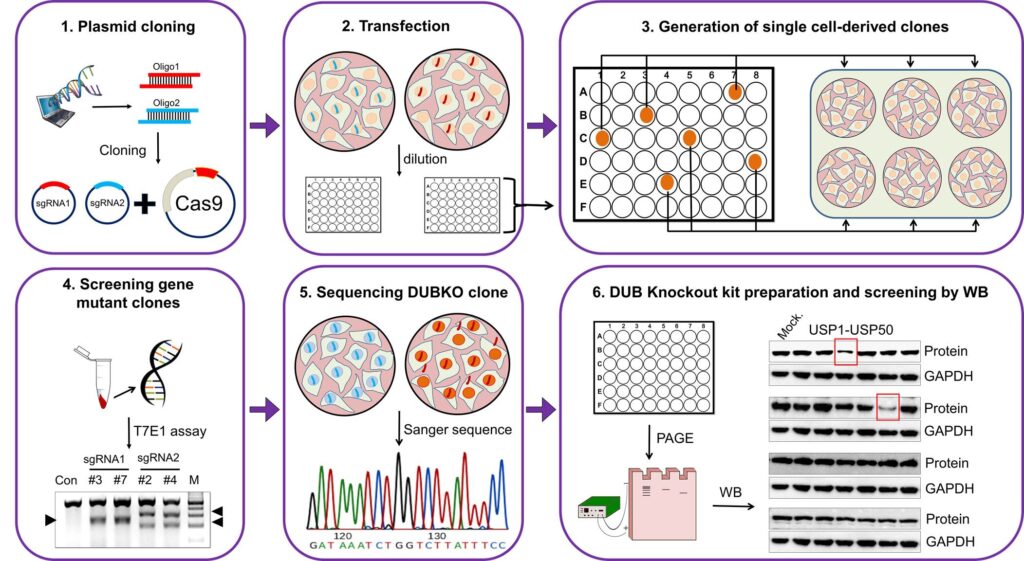
We recently generated a Deubiquitinating enzymes knockout library kit using the CRISPR/Cas9 system to screen ubiquitin-specific proteases (USPs) to identify candidates that regulate protein turnover of any protein of our interest by reversing its ubiquitination, thereby protecting it from degradation. We generated a library of 50 individual knockout cell lines targeting the genes of each USP family member. The cell lysates from these USP knockout cell lines were used in western blot analyses to screen for DUB candidates that regulate target protein stabilization. Our approach can provide new insights into the molecular mechanisms by which DUBs regulate their target proteins in diverse cellular processes and wide range of biomedical fields
